Training Machine Learning Models to Predict Solubility of Molecules
jekyll jupyter_notebook featured machine_learning regression from IPython.core.display import HTML
Building Machine Learning Models in Python
This notebook shows how a machine learning model can be built in python
The steps to accomplish this consist of:
- Get/choose data! --> (Via Toy datasets or through own personal collection)
- Load data onto notebook through pandas and import desired regressor models
- Assign dependent and indenpendent variables from dataset
- Split the data into a training set and a test set
- Choose a regressor (linear regression, random forest or something else)
- Determine the model performance (statistics like MSE and R2)
- Compare against other regressors?
- Visualize results
Table of Contents
- Step 1: Data loading and inspection
- Step 2: Generate Training and Test Sets
- Step 3: Running the Regressor Models
- Step 4: Compare Regressor Performance
- Step 5: Visualize Results
- Full Routines
- Conclusions
The following modules are commonly used in machine learning (ML) applications.
- pandas : Used to load, process, analyze, operate and export dataframes
- sklearn : Contains a variety of modeling and statistical tools useful for ML
- matplotlib.pyplot :Used for plotting/visualizing our results
- seaborn : A powerful plotting utility built off matplotlib
- numpy : Used to perform computations on array data
The API documentation for each of these modules can be found here:
pandas is generally abbreviated as pd , numpy as np, matplotlib.pyplot as plt, and seaborn as sns.
#Import the necessary modules
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression, HuberRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neural_network import MLPRegressor
from sklearn.metrics import mean_squared_error, r2_score
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
Step 1: Data loading and inspection
This section will delineate the basic steps of loading and preliminary inspection of our data using a few pandas methods. If you are faimiliar with pandas you can skip this section :)
Loading data with pandas
The first thing that needs to be done is load the data onto our jupyter notebook.There's a variety of methods to accomplish this.
In this case, a .csv file (Commma Separated Values) containing solubility data for a variety of molecules extracted from a study conducted by John S. Delaney (found here: https://pubs.acs.org/doi/10.1021/ci034243x) will be used.
Loading data from .csv files is accomplished via the read_csv function in pandas.
Here I'll be naming the dataframe from our .csv file as df
#Load the data from the .csv file and see the resulting dataframe
df = pd.read_csv('delaney_solubility_with_descriptors.csv',index_col=False)
df.head()
| MolLogP | MolWt | NumRotatableBonds | AromaticProportion | logS | |
|---|---|---|---|---|---|
| 0 | 2.5954 | 167.850 | 0.0 | 0.0 | -2.18 |
| 1 | 2.3765 | 133.405 | 0.0 | 0.0 | -2.00 |
| 2 | 2.5938 | 167.850 | 1.0 | 0.0 | -1.74 |
| 3 | 2.0289 | 133.405 | 1.0 | 0.0 | -1.48 |
| 4 | 2.9189 | 187.375 | 1.0 | 0.0 | -3.04 |
Inspecting and understanding the loaded data with pandas
I used the '.head()' method to visualize the first 5 rows on the resulting dataframe. You can visualize the first and last 5 rows in your dataframe by simply entering the name of your dataframe. You can also visualize the first 'N' rows in your dataframe by passing an integer value onto .head().
Our dataframe has 5 columns that contain the following information for each molecule:
The $\log P$ value --> $\log P$ is the logarithm of the partition coefficient and represents the ratio of concentrations of a compound in a mixture of two immiscible solvents at equilibrium.
$\log P = \log\left ( \frac{[A]_{solute, organic}}{[A]_{solute, aqueous}} \right ) = \log K_{D}$
In the equation above, $[A]$ represents the concentration of compound A in one of the layers of interest. The ratio of concentrations in two different layers is also known as the partition constant $K_{D}$. The partition coefficient is a quantity regularly used in chemistry to characterize the extent of hydrophilicity of chemical compounds. This quantity is unitless.
The image below shows a density column made up of 5 different liquid compounds. The liquids separate because they have dismilar solubilities between one another.

The MolWt --> This corresponds to the molecular weight which is the sum of the weights of the atoms that make up a molecule. This is typically reported in units of grams/mole [g/mol]
The number of rotatable bonds --> Atoms in molecules are connected via chemical bonds. Bonds come in a variety of different types but the ones we are concerned here are single, double and triple. Single bonds can freely rotate in space while double and triple bonds are rigid. Therefore, this column represents the number of single bonds in the molecule that are not within a ring structure.
The aromatic proportion --> This is the ratio of the number of aromatic atoms in a molecule and the number of heavy atoms in a molecule.
The $\log S$ value --> This is the logarithm of the solubility coefficient (S). In Delaney's study he was interested in determining the solubility of a variety of compounds in aqueous solutions (i.e., water). The more positive this value, the more soluble the compound will be.
It's important to understand the data we are working with since this will aid us as we move forward with any necessary processing, analysis, and subsequent interpretations of said analysis.
Let's find out how many molecules this dataset contains using the '.shape' method in pandas.
mols, cols = df.shape;
print('The Delaney dataset contains solubility data for ' + str(mols) + ' molecules')
The Delaney dataset contains solubility data for 1144 molecules
fig, ax = plt.subplots(2, 5, figsize=(30,15))
#Plot histograms for each column of dataframe
sns.histplot(df, x = "MolLogP" , ax = ax[0,0])
sns.histplot(df, x = "MolWt" , ax = ax[0,1])
sns.histplot(df, x = "NumRotatableBonds" , ax = ax[0,2])
sns.histplot(df, x = "AromaticProportion", ax = ax[0,3])
sns.histplot(df, x = "logS" , ax = ax[0,4])
#Show KDE
sns.kdeplot(data = df, x = "MolLogP" , ax = ax[1,0])
sns.kdeplot(data = df, x = "MolWt" , ax = ax[1,1])
sns.kdeplot(data = df, x = "NumRotatableBonds" , ax = ax[1,2])
sns.kdeplot(data = df, x = "AromaticProportion", ax = ax[1,3])
sns.kdeplot(data = df, x = "logS" , ax = ax[1,4])
plt.show()
minVals = df.min()
maxVals = df.max()
rangeVals = (df.max() - df.min())
medians = df.median()
means = df.mean()
stds = df.std()
df_Props = pd.DataFrame([minVals, maxVals, rangeVals,medians,means,stds]).transpose()
df_Props.columns = ['Min','Max','Range', 'Medians', 'Means','SDs']
#Center align the information, and change the output precision for cleaner display of dataframe output
pd.set_option('precision', 3)
dfStyler = df_Props.style.set_properties(**{'text-align': 'center'})
dfStyler.set_table_styles([dict(selector='th', props=[('text-align', 'center')])])
df_Props = df_Props.style.set_caption('Summary of Descriptive Statistics for Delaney Solubility Data')
df_Props
| Min | Max | Range | Medians | Means | SDs | |
|---|---|---|---|---|---|---|
| MolLogP | -7.571 | 10.389 | 17.960 | 2.340 | 2.449 | 1.866 |
| MolWt | 16.043 | 780.949 | 764.906 | 183.595 | 204.632 | 102.620 |
| NumRotatableBonds | 0.000 | 23.000 | 23.000 | 1.000 | 2.174 | 2.627 |
| AromaticProportion | 0.000 | 1.000 | 1.000 | 0.375 | 0.365 | 0.343 |
| logS | -11.600 | 1.580 | 13.180 | -2.870 | -3.058 | 2.097 |
From the results above we can tell the following:
- The partition coefficients span values between -7.57 to 10.39. The median value is 2.34 and the average value is 2.45. The distribution appears unimodal from the KDE.
- The molecular weights span values between 16.04 to 780.95 g/mol. The median value is 183.59 g/mol and the average value is 102.62 g/mol. The distribution appears approximately bimodal from the KDE.
- The number of rotatable bonds spans values between 0 to 23. The median value is 1 and the average value is 2.63. The distribution appears approximately skewed unimodal from the KDE.
- The aromatic proportions span values between 0 to 1. The median value is 0.38 and the average value is 0.34. The distribution appears bimodal from the KDE.
- The solubility coefficients span values between -11.6 to 1.58. The median value is -2.87 and the average value is -2.87. The distribution appears bimodal from the KDE.
We can also look at the bivariate distribution between the partition coefficient and the solubility coefficient as shown below.
sns.displot(df, x = "MolLogP", y = "logS", cbar = True)
sns.displot(df, x = "MolLogP", y = "logS", kind = "kde")
<seaborn.axisgrid.FacetGrid at 0x22216bfc448>
From the bivariate distribution histrogram shown above uses a color scale (the z-axis corresponding to counts) where observations with low counts are white/light while observations with high counts are shown as dark/dark.
We can see that there is a maxima situated at $\log P$ ~ 3 and $\log S$ ~ -3 which is consistent with the mean values we determined earlier.
Importantly, it can be seen that there appears to be an approximately linear relationship between $\log P$ and $\log S$. $\log S$ seems to be inversely proportional to $\log P$. This is important to note because knowing that these two variables have this relationship can allow us to use a linear regression model to train our dataset.
This can also be readily visualized using the jointplot display shown below.
sns.jointplot(data=df, x="MolLogP", y="logS")
<seaborn.axisgrid.JointGrid at 0x22217398b88>
The same treatment can be applied to all the other parameters in our datasets and is displayed in the pairplot shown below.
The results of these visualizations elucidate that there are a few potentially linear relationships present between parameters that we could explore on our training model (i.e., $\log P$ vs MolW and MolWt vs $\log S$).
sns.pairplot(df)
<seaborn.axisgrid.PairGrid at 0x22217beb448>
g = sns.PairGrid(df)
g.map_upper(sns.histplot)
g.map_lower(sns.kdeplot, fill=True)
g.map_diag(sns.histplot, kde=True)
<seaborn.axisgrid.PairGrid at 0x22219d56608>
The last visualization that I'll show here before jumping into the training of the model is shown below.
This plot takes $\log P$ (independent variable, x-axis), and plots it against $\log S$ (dependent variable, y-axis). The color of each datapoint represents the molecular weight associated with that observation (darker shades mean higher molecular weights). The size of each datapoint is also dictated by the molecular weight (bigger circles mean higher molecular weights).
From this plot it can be readily seen that logS decreases as a function of $\log P$. This means that as the partition coefficient increases, the solubility of the molecule decreases. Furthermore, the data shows that $\log S$ also generally decreases with increasing molecular weight which makes physical sense.
#Plot relationship between partition coefficient and solubility coeffient
sns.relplot(x = "MolLogP", y = "logS", hue = "MolWt", size = "MolWt", sizes=(15, 200), data = df)
<seaborn.axisgrid.FacetGrid at 0x2221c0cc6c8>
Step 2: Generate Training and Test Sets
Now that we understand our data and have taken the time to visualize it, we can start training our machine learning model.
Define the independent and dependent variables
To accomplish this, we need to decide what variable we would like to predict (i.e. our dependent variable), and decide which of our other available parameter(s) will serve as our independent variable(s).
In this case, we will set our independent variable (y) to be logS and the remaining columns will serve as our dependent variables (x).
#Assigning the value for x [independent variables]
x = df.drop(['logS'], axis = 1)
x
| MolLogP | MolWt | NumRotatableBonds | AromaticProportion | |
|---|---|---|---|---|
| 0 | 2.595 | 167.850 | 0.0 | 0.000 |
| 1 | 2.377 | 133.405 | 0.0 | 0.000 |
| 2 | 2.594 | 167.850 | 1.0 | 0.000 |
| 3 | 2.029 | 133.405 | 1.0 | 0.000 |
| 4 | 2.919 | 187.375 | 1.0 | 0.000 |
| ... | ... | ... | ... | ... |
| 1139 | 1.988 | 287.343 | 8.0 | 0.000 |
| 1140 | 3.421 | 286.114 | 2.0 | 0.333 |
| 1141 | 3.610 | 308.333 | 4.0 | 0.696 |
| 1142 | 2.562 | 354.815 | 3.0 | 0.522 |
| 1143 | 2.022 | 179.219 | 1.0 | 0.462 |
1144 rows × 4 columns
#Assigning the value for y [dependent variable]
#y = df.iloc[:,-1]
#y = df.logS
y = df['logS']
y
0 -2.180
1 -2.000
2 -1.740
3 -1.480
4 -3.040
...
1139 1.144
1140 -4.925
1141 -3.893
1142 -3.790
1143 -2.581
Name: logS, Length: 1144, dtype: float64
Splitting the data for model development
Now that we have defined our dependent and independent variables, we can split our data into a training set and a test set.
A training set is a dataset used during the learning process and is used to fit the parameters
A test set is a set of examples used only to assess the performance of the model.
The splitting of the data is an extremely important element of the model building process.
When trying to determine the splitting of the data there are two competing concerns that one needs to consider:
- As the proportion of training data decreases the parameter estimates will exhibit increased variance.
- As the proportion of testing data decreases the assesment of the model performance statistics will have greater variance.
Therefore, when choosing how to split the data we are seeking to minimize the variance associated with each of those concerns. The best splitting ultimately depends on the size of your dataset. The exact splitting of the dataset matters less when the you have a large dataset. At that point, the splitting can allow for smaller training sets to be used in order to expedite computationally intensive processes.
For the work shown here I'll make use of the commonly employed 80/20 rule (also known as the Pareto principle or Pareto splitting). The 80/20 rule basically states that 80% of effects come from 20% of the causes. The 80/20 rule is generally a good starting point for data splitting.
If the 80/20 rule does not seem to produce good predictions on your data then exploring other splittings could be beneficial. This could be done by systematically adjusting the splits and checking the resulting variances in the model or by making use of something like a scaling law.
We split our model using train_test_split function from scikit-learn.
1) X_train - This contains all independent variables which will be used to train the model. In the example here, I've specified the test_size = 0.2, this means 80% of observations from the total/complete data will be used to train/fit the model and remaining 20% will be used to test the model.
2) X_test - This is the remaining 20% portion of the independent variables from the data that will not be used in the training phase. These values will be used to test the accuracy of the model.
3) y_train - This is the dependent variable which needs to be predicted by our model. This includes category labels against your independent variables, we need to specify our dependent variable while training/fitting the model.
4) y_test - This data has category labels for the test data which will be used to test the accuracy between the actual and predicted values.
#Data splitting for model development
#Define test set as consisting of 20% of the data while 80% of the data goes to the training set
#Training set: subset to train a model
#Test set: subset to test the trained model
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = 0.2, random_state = 42)
Different Types of Regressor Modelst
Regressor models fall into the following categories:
- Ensemble Methods
- Classical/Linear Methods
- Gaussian Process Methods
- Bayesian Methods
- Variable Selection Methods
- Outlier-robust Methods
- Generalized linear models Methods
- Miscellaneous Methods
- Nearest Neighbors Methods
- Neural Networks Methods
- Decision Tree Methods
- Composite Estimator Methods
- Cross decomposition Methods
In this document I'll explore models trained via 6 different methods to see how well they predict the solubility from the Delaney data. The methods I'll explore are:
- Linear Method (Linear Regression)
- Ensemble Method (Random Forest)
- Gaussian Process (Gaussian Process)
- Outlier-robust (Huber Regressor)
- Nearest Neighbors (K-Nearest Neighbors)
- Neural Network (MLP)
For each of these models I'll provide a brief description and introduction as to how they work. Then, I'll show how the model is set up for training followed by an assessment of their performance using the $r^{2}$ score and the mean squared error (MSE) obtained by comparing the solubility values that we partitioned into the training set and the solubility values that our regression model predicted.
Step 3: Running the Regressor Models
The following sections will describe show the setup and performance assessment of each of the regression models to be utilized for solubility prediction.
Linear Regression Basics
Now that we have split the data into test and training sets, we can decide what model to use. In this example, a linear regression will be performed. Linear regression is the process of measuring the output from a set of inputs. The simplest linear regression model is a line expressed mathematically as:
$\hat{Y} = \hat{\beta}_{0} + \sum \limits _{j=1} ^{p} X_{j}\hat{\beta}_{j} $
In this equation $\hat{\beta}_{0}$ and $\hat{\beta}_{j}$ are the fit parameters of the model (the y intercept and the slope of the line in this case).
If our independent variable consists of a single input then this a simple linear regression. However, we are usually interested in the effect that many variables (i.e., inputs) have on our dependent variable. For this kind of analysis we need to use a technique like Ordinary Least Squares, Gradient Descent, or Regularization. Each of these techniques seeks to minimize a different quantity.
In this example, we'll be using the Ordinary Sum of Squares (OSS). In OSS, the residual sum of squares (RSS) is what is being minimized. The RSS is expressed as:
$RSS = \sum_{i}^{N} \left ( y_{i} - f(x_{i})\right)^{2}$
Here $y_{i}$ is the measured value and $f(x_{i})$ is the model value. The lower the RSS the more accurate the model.
I also wanted to briefly talk about regularization methods. Regularization methods are useful because they aim to minimize the RSS while simultaneously reducing the complexity of the model (i.e, minimizing the potential of overfitting). The reduction in the complexity of a model is important to take into account because a model with an arbitrarily large number of parameters begins to suffer from the uniqueness problem (i.e., other parameter combinations can also produce good models/fits for our data).
Two common ways to address the complexity/over-fitting concerns are via L1 and L2 regularization. These regularization techniques introduce a penalty term to the loss/cost function associated with the model. In L1 regularization the cost function (CF) looks like this:
$CF_{L2} = \sum_{i}^{N} (y_{i} - x_{ij}\beta_{j})^{2} +\lambda \sum_{j}^{N} \beta_{j}^{2}$
Here $y_{i}$ are the observed values, $ x_{ij}\beta_{j}$ are the model values, $\beta_{j}$ is a weight term, and $\lambda$ is known as the regularization rate.
In L2 regularization the cost function (CF) looks like this:
$CF_{L1} = \sum_{i}^{N} (y_{i} - x_{ij}\beta_{j}))^{2} +\lambda \sum_{j}^{N} \left | \beta_{j} \right |$
As you can see, these two approaches are identical except for the second term which is known as the penalty term. These differences are summarized in the following points:
- L1 is robust to outliers
- L1 has built-in feature selection
- L1 is a sparse solution (i.e., regularizes has many 0 values)
- L1 penalizes the sum of the absolute weights
Initiating and Running a Linear Regression
#Perform linear regression to build the model
lr = LinearRegression() #Initiate linear regression variable
lr.fit(X_train, y_train) #Train the model
y_lr_train_pred = lr.predict(X_train) #Predicted y values using training set
y_lr_test_pred = lr.predict(X_test) #Predicted y values using test set
Assessing the Performance of Linear Regression Model
#Determine performance of linear regression model through mean squared error (MSE) and R2 on training/test sets
lr_train_mse = mean_squared_error(y_train, y_lr_train_pred)
lr_train_r2 = r2_score(y_train, y_lr_train_pred)
lr_test_mse = mean_squared_error(y_test, y_lr_test_pred)
lr_test_r2 = r2_score(y_test, y_lr_test_pred)
#Make pandas dataframe that contains results of model training
lr_results = pd.DataFrame(['Linear regression',lr_train_mse, lr_train_r2, lr_test_mse, lr_test_r2]).transpose()
lr_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
print('The MSE for the training set is:')
print(lr_train_mse)
print('The MSE for the test set is:')
print(lr_test_mse)
lr_results
The MSE for the training set is: 1.0139894491573003 The MSE for the test set is: 0.9990844407075308
| Method | Training MSE | Training R2 | Test MSE | Test R2 | |
|---|---|---|---|---|---|
| 0 | Linear regression | 1.014 | 0.77 | 0.999 | 0.771 |
The results of the model are summarized above.
The $r^{2}$ is 0.77 which means this model has an accuracy score of 77%. For a 'perfect; fit, $r^{2}$ = 1 and indicates that the dependent and independent variables are highly correlated. An $r^{2}$ above 0.9 is generally desired. The $r^{2}$ obtained is rather low and suggests that the linear regression model is not fitting/representing the data too well.
The MSE of the training set is 1.01. The MSE is a quantity that needs to be minimized. For a 'perfect' model, there would be no errors between the data and the model and the MSE would be 0. Notice how the MSE of the training set is HIGHER than the MSE of the test set. This is not a good sign because the point of the training set is to optimize for a low MSE whereas your test set MSE is calculated over data unseen during the optimization.
Random Forest Basics
Random Forest (RF) is a supervised learning algorithm based on ensemble learning method (i.e., combine predictions from multiple machine learning models) and decision trees. All calculations are run in parallel which means that there is no interaction between the decision trees as they are built. RF is often used to solve classification and regression problems.
The schematic below shows how RF is structured

The RF algorithm works as follows:
- Pick $k$ data points from the training set at random
- Build decision tree from those $k$ points
- Select number $N$ of decision trees to build
- [Optional] Select how deep into each forest to go
- Repeat steps 1 and 2
- Use each of the $N$ trees to predict the value of each data point and use the new data point to the average of all predicted values
Initiating and Running a Random Forest
The code setup for a RF regression is essentially identical to setting up a linear regression. The main difference is that for the RF model, we can specify other parameters such as the 'max_depth' parameter which indicates how deep (i.e. how many 'splits') into the decision tree to go.
#Perform random forest to build the model
rf = RandomForestRegressor(max_depth=3, random_state=42)
rf.fit(X_train, y_train)
#Predicted values of training and test sets for random forest
y_rf_train_pred = rf.predict(X_train)
y_rf_test_pred = rf.predict(X_test)
#Determine performance of random forest
rf_train_mse = mean_squared_error(y_train, y_rf_train_pred)
rf_train_r2 = r2_score(y_train, y_rf_train_pred)
rf_test_mse = mean_squared_error(y_test, y_rf_test_pred)
rf_test_r2 = r2_score(y_test, y_rf_test_pred)
#Store results of random forest
rf_results = pd.DataFrame(['Random forest',rf_train_mse, rf_train_r2, rf_test_mse, rf_test_r2]).transpose()
rf_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
rf_results
| Method | Training MSE | Training R2 | Test MSE | Test R2 | |
|---|---|---|---|---|---|
| 0 | Random forest | 0.751 | 0.829 | 0.814 | 0.813 |
Assessing the Performance of the Random Forest Model
The results of the model are summarized above.
The $r^{2}$ is 0.83 which gives it an accuracy score of 83%. The $r^{2}$ obtained is still a bit low and suggests that the random forest regression model is still not fitting/representing the data too well.
The MSE of the training set is 0.75. In this case, the MSE of the training set is lower than the MSE of the test set. Therefore, the random forest regressor is indeed improving the fit quality.
Gaussian Process Basics
The Gaussian Process model is a probabilistic supervised machine learning framework that is widely used for regression and classification. A Gaussian processes regression (GPR) model can make predictions incorporating prior knowledge (kernels) and provide uncertainty measures over predictions. Here's a wonderful article that covers Gaussian Processes in detail. I'll just be summarizing the key points here.
A random variable X is said to be Gaussian (aka normally distributed) via the following probability density function (PDF):
$P_{X}(x) = \frac{1}{\sqrt{2\pi}\sigma}e^{\frac{-({x-\mu})^{2}}{2\sigma^{2}}}$
Here, the mean is $\mu$ and the variance is $\sigma$. This distribution looks like the familiar "bell curve".
Each of the points in a Gaussian distribution can be represented as a vector. However, for the univariate case described just now, the resulting functions are not smooth enough to be used for regression tasks. To aid in the smoothing of these functions a joint distribution is needed which can be obtained via multivariate distribution theory and the usage of covariance functions. A special type of covariance functions are kernel functions. Kernel functions are defined by their usuage of inner products (i.e., a generalized form of the dot product). For Gaussian Processes the most commonly used kernel function is the squared exponential which looks like this:
$cov(x_{i},x_{j}) = e^{\frac{-({x_{i}-x_{j}})^{2}}{2}}$
Here $x_{i}$ and $x_{j}$ are values derived from the input vectors I mentioned earlier.
In a Gaussian Process model, we generate a probability distribution based on functions that fit a set of points. Since we have this probability distribution, we can also determine the means and variance of our resulting model at each iteration.
Initiating and Running a Gaussian Process Regression
The code setup for a Gaussian Process regression is similar to what we've done before.
#Perform Gaussian Process to build the model
gp = GaussianProcessRegressor(random_state=42)
gp.fit(X_train, y_train)
#Predicted values of training and test sets for Gaussian Process
y_gp_train_pred = gp.predict(X_train)
y_gp_test_pred = gp.predict(X_test)
#Determine performance of Gaussian Process
gp_train_mse = mean_squared_error(y_train, y_gp_train_pred)
gp_train_r2 = r2_score(y_train, y_gp_train_pred)
gp_test_mse = mean_squared_error(y_test, y_gp_test_pred)
gp_test_r2 = r2_score(y_test, y_gp_test_pred)
#Store results of random forest
gp_results = pd.DataFrame(['Gaussian Process',gp_train_mse, gp_train_r2, gp_test_mse, gp_test_r2]).transpose()
gp_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
gp_results
| Method | Training MSE | Training R2 | Test MSE | Test R2 | |
|---|---|---|---|---|---|
| 0 | Gaussian Process | 0.021 | 0.995 | 1066.374 | -243.888 |
Assessing the Performance of the Gaussian Process Model
The results of the model are summarized above.
The $r^{2}$ is 1.00. The $r^{2}$ obtained indicates a perfect correlation between out variables and suggests that the Gaussian Process regression model is fitting/representing the data really well. This is further evidenced by the massive difference in the MSE of the test set vs training set. Notice that the training set has a much lower MSE than the test MSE which means this regressor is doing a fantastic job!
The MSE of the training set is 0.02 and is SIGNIFICANTLY lower than the MSE of the test set. This means that this regressor results in an excellent fit.
Huber Regression Basics
Huber Regressor is a regression technique that is robust to outliers. The idea is to use a loss function that has been modified from the standard least-squares method. The loss function used in Huber regression is similar to the least squares penalty for small residuals. However, when the residuals are large the penalty is lower and increases linearly rather than quadratically. This is what makes Huber Regression more forgiving of outliers in our data. Whether a residual is considered small or large is based on a threshold value denoted as $\epsilon$
The cost function for the Huber method is defined as follows:
$CF_{HR} = \sum_{i=1}^{m} \phi(y_{i} -x_{i}^{T}\beta )$
The Huber function for the Huber method is defined as follows:
$\phi = \begin{cases} u^{2} & \left | u \right | \leq \epsilon\\ 2\epsilon u -\epsilon ^{2} & \left | u \right | > \epsilon \\ \end{cases} $
The distinguishing factor of the Huber method is $\epsilon$. As $\epsilon$ gets smaller the HR method becomes more robust to outliers.
Reference: Huber, P. J. 1964. “Robust Estimation of a Location Parameter.” Annals of Mathematical Statistics 35 (1): 73–101.
Initiating and Running a Huber Regressor
#Perform Huber Regressor to build the model
hr = HuberRegressor(epsilon=1.01)
hr.fit(X_train, y_train)
#Predicted values of training and test sets for Huber Regressor
y_hr_train_pred = hr.predict(X_train)
y_hr_test_pred = hr.predict(X_test)
#Determine performance of Huber Regressor
hr_train_mse = mean_squared_error(y_train, y_hr_train_pred)
hr_train_r2 = r2_score(y_train, y_hr_train_pred)
hr_test_mse = mean_squared_error(y_test, y_hr_test_pred)
hr_test_r2 = r2_score(y_test, y_hr_test_pred)
#Store results of Huber Regressor
hr_results = pd.DataFrame(['Huber Regressor',hr_train_mse, hr_train_r2, hr_test_mse, hr_test_r2]).transpose()
hr_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
hr_results
| Method | Training MSE | Training R2 | Test MSE | Test R2 | |
|---|---|---|---|---|---|
| 0 | Huber Regressor | 1.028 | 0.766 | 1.029 | 0.764 |
Assessing the Performance of the Huber Regressor Model
The results of the model are summarized above.
The $r^{2}$ is 0.77. The $r^{2}$ obtained is rather low and suggests that the Huber Regressor model is not fitting/representing the data well. The training set has a slightly lower MSE than the test MSE but the difference is insignificant.
K-Nearest Neighbors Regression Basics
K-Nearest Neighbors is a non-parametric supervised learning method. The KNN algorithm uses ‘feature similarity’ to predict the values of any new data points. This means that the new point is assigned a value based on how closely it resembles the points in the training set. KNN algorithm can be used for both classification and regression problems. Here's a nice animation I found that showcases the basic principles of KNN:
Initiating and Running a Nearest Neighbors Regression
The code setup for a Nearest Neighbors Regressor is similar to the regressions we've already done. The main difference is that we can specify a 'n_neighbors' value that controls number of nearest neighbors to consider as part of the process.#Perform Nearest Neighbors Regression to build the model
kn = KNeighborsRegressor(n_neighbors=1)
kn.fit(X_train, y_train)
#Predicted values of training and test sets for Nearest Neighbors Regression
y_kn_train_pred = kn.predict(X_train)
y_kn_test_pred = kn.predict(X_test)
#Determine performance of Nearest Neighbors Regression
kn_train_mse = mean_squared_error(y_train, y_kn_train_pred)
kn_train_r2 = r2_score(y_train, y_kn_train_pred)
kn_test_mse = mean_squared_error(y_test, y_kn_test_pred)
kn_test_r2 = r2_score(y_test, y_kn_test_pred)
#Store results of Nearest Neighbors Regression
kn_results = pd.DataFrame(['Nearest Neighbors',kn_train_mse, kn_train_r2, kn_test_mse, kn_test_r2]).transpose()
kn_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
kn_results
| Method | Training MSE | Training R2 | Test MSE | Test R2 | |
|---|---|---|---|---|---|
| 0 | Nearest Neighbors | 0.031 | 0.993 | 1.523 | 0.65 |
Assessing the Performance of K-Nearest Neighbors Regressor Model
We've succesfully ran the Huber Regressor model on our test and training sets derived from our data. In order to determine how well the models performed we'll once again use the MSE and $r^{2}$ coefficient.
The results of the model are summarized above.
The $r^{2}$ is 0.99. The $r^{2}$ obtained is quite high and suggests that the K-Nearest Neighbors Regression model is fitting/representing the data quite well. The training set a significantly lower MSE than the test MSE which means that the KNN is optimizing our model excellently.
Neural Network Regression Basics
Neural Networks are one of the deep learning algorithms that simulate the workings of neurons in the human brain. The purpose of using Neural Networks for Regression is that these models can learn non-linear relationship betweens the features and the target and therefore can model more complex relationships. Neural Networks have the ability to learn the complex relationship between the features and target due to the presence of activation functions.
Neural Networks consist of 3 main layers, namely an Input layer, a Hidden layer, and an Output layer. Each layer consists of $N$ neurons. Each layer will have an Activation Function associated with each of the neurons. The activation function is responsible for introducing non-linearity in the model. Each layer can also have regularizers which aid in the prevention of overfitting.
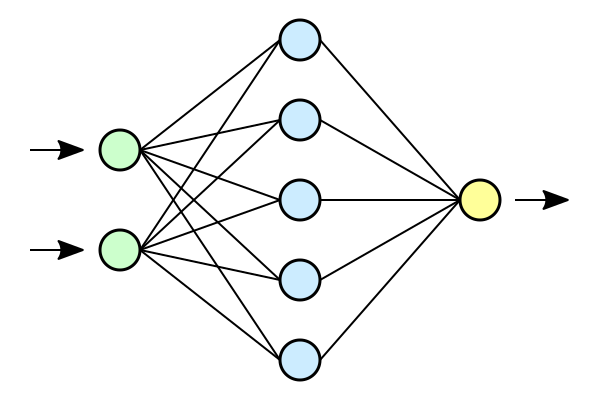
Initiating and Running a Neural network Regression
The code setup for a Neural network Regressor is similar to the regressions we've already done. However, there are more parameters at our disposal that we can play with like the choice of solver, number of hidden layers, and the learning rate amongst others.
#Perform Neural network Regression to build the model
nn = MLPRegressor(hidden_layer_sizes = 500, solver='adam', learning_rate_init = 1e-2, shuffle = True,
random_state = 42)
nn.fit(X_train, y_train)
#Predicted values of training and test sets for Neural network Regression
y_nn_train_pred = nn.predict(X_train)
y_nn_test_pred = nn.predict(X_test)
#Determine performance of Neural network Regression
nn_train_mse = mean_squared_error(y_train, y_nn_train_pred)
nn_train_r2 = r2_score(y_train, y_nn_train_pred)
nn_test_mse = mean_squared_error(y_test, y_nn_test_pred)
nn_test_r2 = r2_score(y_test, y_nn_test_pred)
#Store results of Neural network Regression
nn_results = pd.DataFrame(['Neural network',nn_train_mse, nn_train_r2, nn_test_mse, nn_test_r2]).transpose()
nn_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
nn_results
| Method | Training MSE | Training R2 | Test MSE | Test R2 | |
|---|---|---|---|---|---|
| 0 | Neural network | 0.744 | 0.831 | 0.663 | 0.848 |
Assessing the Performance of MLP Neural Network Regressor Model
The results of the model are summarized above.
The $r^{2}$ is 0.99. The $r^{2}$ obtained is quite high and suggests that the K-Nearest Neighbors Regression model is fitting/representing the data quite well. The training set a significantly lower MSE than the test MSE which means that the KNN is optimizing our model excellently.
Comparing the Performance of the Different Models
The performance results from all 6 models explored here are shown in the table below. The table has been soted so that the model with the highest $r^{2}$ value is at the top. As can be seen, the Gaussian Process and K-Nearest Neighbors produced the models with the best $r^{2}$ values and lowest MSE. The linear regression and Huber regression models performed the worst.
#Combine results from all our models into a single dataframe
mod_Results = pd.concat([lr_results, rf_results,gp_results,hr_results,kn_results,nn_results],
ignore_index=True).sort_values('Training R2',axis = 0, ascending = False)
mod_Results
| Method | Training MSE | Training R2 | Test MSE | Test R2 | |
|---|---|---|---|---|---|
| 2 | Gaussian Process | 0.021 | 0.995 | 1066.374 | -243.888 |
| 4 | Nearest Neighbors | 0.031 | 0.993 | 1.523 | 0.65 |
| 5 | Neural network | 0.744 | 0.831 | 0.663 | 0.848 |
| 1 | Random forest | 0.751 | 0.829 | 0.814 | 0.813 |
| 0 | Linear regression | 1.014 | 0.77 | 0.999 | 0.771 |
| 3 | Huber Regressor | 1.028 | 0.766 | 1.029 | 0.764 |
#I'm going to put the plotting routines from above into a loop
listOfPredYVals = [y_lr_train_pred, y_rf_train_pred, y_gp_train_pred,
y_hr_train_pred, y_kn_train_pred, y_nn_train_pred]
hexColorList = ["#415c90", "#ac4228", "#28812e", "#581845", "#FF5733", "#FFC300"]
labelList = ["LR","RF","GP","HR","KNN","NN"]
titleList = ['Linear Regression Result','Random Forest Result',
'Gaussian Process Result', 'Huber Process Result',
'K-Nearest Neighbors Result', 'Neural Network Result']
#Visualize results of linear regression
plt.rcParams.update({'font.size': 20})
pltRows = 3 #
pltCols = 2 #
fig, axs = plt.subplots(nrows = pltRows, ncols = pltCols, figsize=(15,15))
i = 0; j = 0; k = 0;
for element in listOfPredYVals:
color = hexColorList[k]
yTrainPredPlot = listOfPredYVals[k]
label = labelList[k]
#Make scatter plot of train set and regressor model
axs[i,j].scatter(x = y_train, y = yTrainPredPlot, c=color, alpha=0.5)
#Fit a first order polynomial (i.e. a straight line) to the regressor model
z = np.polyfit(y_train, yTrainPredPlot, 1)
p = np.poly1d(z)
#Add labels and colors and stuff
val = mod_Results.loc[k,'Training R2'] #Get the r2 value from the model results dataframe
val = "{:.2f}".format(val)
axs[i,j].plot(y_train,p(y_train),"#b20cd7", label=label+"\nr\u00b2".format(2) + " = " + str(val))
axs[i,j].title.set_text(titleList[k])
k += 1
if i < pltRows-1:
i += 1
else:
i = 0
if j < pltCols-1:
j += 1
else:
j = 0
k = 0
for ax in axs.flat:
ax.set(xlabel='Experimental LogS', ylabel='Predicted LogS')
ax.label_outer()
ax.legend(loc="upper left")
ax.grid(color = 'black', linestyle = '--', linewidth = 0.5)
plt.show()
6 regression models were explored here, however, scikit-learn has a multitude of options for regression available, some of which are listed below:
List of regressors:
- sklearn.linear_model.Ridge
- sklearn.linear_model.SGDRegressor
- sklearn.ensemble.ExtraTreesRegressor
- sklearn.ensemble.GradientBoostingRegressor
- - sklearn.tree.DecisionTreeRegressor
- sklearn.tree.ExtraTreeRegressor
- sklearn.svm.LinearSVR
- sklearn.svm.SVR
sklearn API Docs
https://scikit-learn.org/stable/modules/classes.htmlComparing multiple models is really important when training machine learning models. In addition to that, exploring how different input parameters influences the fit quality is important to explore as well. If you are interested in ensemble learning, then creating a variety of models is necessary since that approach takes the results from each of those models and tries to make a better one from those results.
Full Routines
The full routines for the work done here are shown below. The routines shown here can readily be modified to use other regression models and refactored as desired to control even more aspects of the modeling and use for subsequent/additional model comparisons. There's also a wrapper function that will allow the user to specify which regressors they want to run and produce the output I showed in the last section.#Import the necessary modules
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.ensemble import RandomForestRegressor
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
path2csv = 'delaney_solubility_with_descriptors.csv'
#Load the csv file
df = pd.read_csv(path2csv,index_col=False)
#This function trains a linear regression model from a pandas dataframe
def ML_LRModelTrain(df,tSize,indCol,plotResults=True):
"""
Args:
df: (df) Pandas dataframe containing data for training
tSize: (int) Value indicating splitting of data into test and train sets.
Must be between 0 and 1.
indCol (str) Name of column in dataframe that contains the independent variable
Returns:
bbox_rgb: (ndarray, size (y, x, 3)) cropped bounding box
RGB channels
"""
#Assign the value for independent variables
x = df.drop([indCol], axis = 1)
#Assign the value for dependent variable
y = df[indCol]
#Split data into test and training sets
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = tSize, random_state = 42)
#Perform linear regression to build the model
lr = LinearRegression() #Initiate linear regression variable
lr.fit(X_train, y_train) #Train the model
y_lr_train_pred = lr.predict(X_train) #Predicted y values using training set
y_lr_test_pred = lr.predict(X_test) #Predicted y values using test set
#Determine performance of linear regression model through mean squared error (MSE) and R2 on training/test sets
lr_train_mse = mean_squared_error(y_train, y_lr_train_pred)
lr_train_r2 = r2_score(y_train, y_lr_train_pred)
lr_test_mse = mean_squared_error(y_test, y_lr_test_pred)
lr_test_r2 = r2_score(y_test, y_lr_test_pred)
#Make pandas dataframe that contains results of model training
lr_results = pd.DataFrame(['Linear regression',lr_train_mse, lr_train_r2, lr_test_mse, lr_test_r2]).transpose()
lr_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
print('The MSE for the training set is:')
print(lr_train_mse)
print('The MSE for the test set is:')
print(lr_test_mse)
#Visualize results of linear regression
if plotResults == True:
plt.rcParams.update({'font.size': 30})
fig, ax = plt.subplots(figsize=(10,10))
plt.scatter(x = y_train, y = y_lr_train_pred, c="#415c90", alpha=0.5)
z = np.polyfit(y_train, y_lr_train_pred, 1)
p = np.poly1d(z)
plt.plot(y_train,p(y_train),"#b20cd7", label="LR")
ax.set(xlabel='Observed ' + indCol, ylabel='Predicted ' + indCol)
plt.legend(loc="upper left")
ax.title.set_text('Linear Regression Result')
return y_lr_train_pred, lr_results
#This function trains a random forest model from a pandas dataframe
def ML_RFModelTrain(df,tSize,indCol,plotResults=True):
"""
Args:
df: (df) Pandas dataframe containing data for training
tSize: (int) Value indicating splitting of data into test and train sets.
Must be between 0 and 1.
indCol (str) Name of column in dataframe that contains the independent variable
Returns:
bbox_rgb: (ndarray, size (y, x, 3)) cropped bounding box
RGB channels
"""
#Assign the value for independent variables
x = df.drop([indCol], axis = 1)
#Assign the value for dependent variable
y = df[indCol]
#Split data into test and training sets
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = tSize, random_state = 42)
#Perform random forest to build the model
rf = RandomForestRegressor(max_depth=2, random_state=42)
rf.fit(X_train, y_train)
#Predicted values of training and test sets for random forest
y_rf_train_pred = rf.predict(X_train)
y_rf_test_pred = rf.predict(X_test)
#Determine performance of random forest
rf_train_mse = mean_squared_error(y_train, y_rf_train_pred)
rf_train_r2 = r2_score(y_train, y_rf_train_pred)
rf_test_mse = mean_squared_error(y_test, y_rf_test_pred)
rf_test_r2 = r2_score(y_test, y_rf_test_pred)
#Store results of random forest
rf_results = pd.DataFrame(['Random forest',rf_train_mse, rf_train_r2, rf_test_mse, rf_test_r2]).transpose()
rf_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
print('The MSE for the training set is:')
print(rf_train_mse)
print('The MSE for the test set is:')
print(rf_test_mse)
#Visualize results of random forest
if plotResults == True:
plt.rcParams.update({'font.size': 30})
fig, ax = plt.subplots(figsize=(10,10))
plt.scatter(x = y_train, y = y_rf_train_pred, c="#ac4228", alpha=0.5)
z = np.polyfit(y_train, y_rf_train_pred, 1)
p = np.poly1d(z)
plt.plot(y_train,p(y_train),"#2c8c2b", label="RF")
ax.set(xlabel='Observed ' + indCol, ylabel='Predicted ' + indCol)
plt.legend(loc="upper left")
ax.title.set_text('Random Forest Result')
return y_rf_train_pred, rf_results
#This function trains a Gaussian Process model from a pandas dataframe
def ML_GPModelTrain(df,tSize,indCol,plotResults=True):
"""
Args:
df: (df) Pandas dataframe containing data for training
tSize: (int) Value indicating splitting of data into test and train sets.
Must be between 0 and 1.
indCol (str) Name of column in dataframe that contains the independent variable
Returns:
bbox_rgb: (ndarray, size (y, x, 3)) cropped bounding box
RGB channels
"""
#Assign the value for independent variables
x = df.drop([indCol], axis = 1)
#Assign the value for dependent variable
y = df[indCol]
#Split data into test and training sets
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = tSize, random_state = 42)
#Perform Gaussian Process to build the model
gp = GaussianProcessRegressor(random_state=42)
gp.fit(X_train, y_train)
#Predicted values of training and test sets for Gaussian Process
y_gp_train_pred = gp.predict(X_train)
y_gp_test_pred = gp.predict(X_test)
#Determine performance of Gaussian Process
gp_train_mse = mean_squared_error(y_train, y_gp_train_pred)
gp_train_r2 = r2_score(y_train, y_gp_train_pred)
gp_test_mse = mean_squared_error(y_test, y_gp_test_pred)
gp_test_r2 = r2_score(y_test, y_gp_test_pred)
#Store results of random forest
gp_results = pd.DataFrame(['Gaussian Process',gp_train_mse, gp_train_r2, gp_test_mse, gp_test_r2]).transpose()
gp_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
print('The MSE for the training set is:')
print(gp_train_mse)
print('The MSE for the test set is:')
print(gp_test_mse)
#Visualize results of Gaussian Process
if plotResults == True:
plt.rcParams.update({'font.size': 30})
fig, ax = plt.subplots(figsize=(10,10))
plt.scatter(x = y_train, y = y_gp_train_pred, c="#ac4228", alpha=0.5)
z = np.polyfit(y_train, y_gp_train_pred, 1)
p = np.poly1d(z)
plt.plot(y_train,p(y_train),"#2c8c2b", label="RF")
ax.set(xlabel='Observed ' + indCol, ylabel='Predicted ' + indCol)
plt.legend(loc="upper left")
ax.title.set_text('Gaussian Process Result')
return y_gp_train_pred, gp_results
#This function trains a Huber Process model from a pandas dataframe
def ML_HPModelTrain(df,tSize,indCol,plotResults=True):
"""
Args:
df: (df) Pandas dataframe containing data for training
tSize: (int) Value indicating splitting of data into test and train sets.
Must be between 0 and 1.
indCol (str) Name of column in dataframe that contains the independent variable
Returns:
bbox_rgb: (ndarray, size (y, x, 3)) cropped bounding box
RGB channels
"""
#Assign the value for independent variables
x = df.drop([indCol], axis = 1)
#Assign the value for dependent variable
y = df[indCol]
#Split data into test and training sets
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = tSize, random_state = 42)
#Initiate Huber Regressor
hr = HuberRegressor(epsilon=1.01)
hr.fit(X_train, y_train)
#Predicted values of training and test sets for Huber Regressor
y_hr_train_pred = hr.predict(X_train)
y_hr_test_pred = hr.predict(X_test)
#Determine performance of Huber Regressor
hr_train_mse = mean_squared_error(y_train, y_hr_train_pred)
hr_train_r2 = r2_score(y_train, y_hr_train_pred)
hr_test_mse = mean_squared_error(y_test, y_hr_test_pred)
hr_test_r2 = r2_score(y_test, y_hr_test_pred)
#Store results of Huber Regressor
hr_results = pd.DataFrame(['Huber Regressor',hr_train_mse, hr_train_r2, hr_test_mse, hr_test_r2]).transpose()
hr_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
print('The MSE for the training set is:')
print(hr_train_mse)
print('The MSE for the test set is:')
print(hr_test_mse)
#Visualize results of Huber Process
if plotResults == True:
plt.rcParams.update({'font.size': 30})
fig, ax = plt.subplots(figsize=(10,10))
plt.scatter(x = y_train, y = y_hr_train_pred, c="#ac4228", alpha=0.5)
z = np.polyfit(y_train, y_hr_train_pred, 1)
p = np.poly1d(z)
plt.plot(y_train,p(y_train),"#2c8c2b", label="RF")
ax.set(xlabel='Observed ' + indCol, ylabel='Predicted ' + indCol)
plt.legend(loc="upper left")
ax.title.set_text('Huber Process Result')
return y_hr_train_pred, hr_results
#This function trains a K-Nearest Neighbors model from a pandas dataframe
def ML_KNNModelTrain(df,tSize,indCol,plotResults=True):
"""
Args:
df: (df) Pandas dataframe containing data for training
tSize: (int) Value indicating splitting of data into test and train sets.
Must be between 0 and 1.
indCol (str) Name of column in dataframe that contains the independent variable
Returns:
bbox_rgb: (ndarray, size (y, x, 3)) cropped bounding box
RGB channels
"""
#Assign the value for independent variables
x = df.drop([indCol], axis = 1)
#Assign the value for dependent variable
y = df[indCol]
#Split data into test and training sets
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = tSize, random_state = 42)
#Perform Nearest Neighbors Regression to build the model
kn = KNeighborsRegressor(n_neighbors=1)
kn.fit(X_train, y_train)
#Predicted values of training and test sets for Nearest Neighbors Regression
y_kn_train_pred = kn.predict(X_train)
y_kn_test_pred = kn.predict(X_test)
#Determine performance of Nearest Neighbors Regression
kn_train_mse = mean_squared_error(y_train, y_kn_train_pred)
kn_train_r2 = r2_score(y_train, y_kn_train_pred)
kn_test_mse = mean_squared_error(y_test, y_kn_test_pred)
kn_test_r2 = r2_score(y_test, y_kn_test_pred)
#Store results of Nearest Neighbors Regression
kn_results = pd.DataFrame(['Nearest Neighbors',kn_train_mse, kn_train_r2, kn_test_mse, kn_test_r2]).transpose()
kn_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
print('The MSE for the training set is:')
print(kn_train_mse)
print('The MSE for the test set is:')
print(kn_test_mse)
#Visualize results of random forest
if plotResults == True:
plt.rcParams.update({'font.size': 30})
fig, ax = plt.subplots(figsize=(10,10))
plt.scatter(x = y_train, y = y_kn_train_pred, c="#ac4228", alpha=0.5)
z = np.polyfit(y_train, y_kn_train_pred, 1)
p = np.poly1d(z)
plt.plot(y_train,p(y_train),"#2c8c2b", label="RF")
ax.set(xlabel='Observed ' + indCol, ylabel='Predicted ' + indCol)
plt.legend(loc="upper left")
ax.title.set_text('K-Nearest Neighbors Result')
return y_kn_train_pred,kn_results
#This function trains a Neural Network model from a pandas dataframe
def ML_NNModelTrain(df,tSize,indCol,plotResults=True):
"""
Args:
df: (df) Pandas dataframe containing data for training
tSize: (int) Value indicating splitting of data into test and train sets.
Must be between 0 and 1.
indCol (str) Name of column in dataframe that contains the independent variable
Returns:
bbox_rgb: (ndarray, size (y, x, 3)) cropped bounding box
RGB channels
"""
#Assign the value for independent variables
x = df.drop([indCol], axis = 1)
#Assign the value for dependent variable
y = df[indCol]
#Split data into test and training sets
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = tSize, random_state = 42)
#Perform Neural network Regression to build the model
nn = MLPRegressor(hidden_layer_sizes = 500, solver='adam', learning_rate_init = 1e-2, shuffle = True,
random_state = 42)
nn.fit(X_train, y_train)
#Predicted values of training and test sets for Neural network Regression
y_nn_train_pred = nn.predict(X_train)
y_nn_test_pred = nn.predict(X_test)
#Determine performance of Neural network Regression
nn_train_mse = mean_squared_error(y_train, y_nn_train_pred)
nn_train_r2 = r2_score(y_train, y_nn_train_pred)
nn_test_mse = mean_squared_error(y_test, y_nn_test_pred)
nn_test_r2 = r2_score(y_test, y_nn_test_pred)
#Store results of Neural network Regression
nn_results = pd.DataFrame(['Neural network',nn_train_mse, nn_train_r2, nn_test_mse, nn_test_r2]).transpose()
nn_results.columns = ['Method','Training MSE','Training R2','Test MSE','Test R2']
print('The MSE for the training set is:')
print(nn_train_mse)
print('The MSE for the test set is:')
print(nn_test_mse)
#Visualize results of random forest
if plotResults == True:
plt.rcParams.update({'font.size': 30})
fig, ax = plt.subplots(figsize=(10,10))
plt.scatter(x = y_train, y = y_nn_train_pred, c="#ac4228", alpha=0.5)
z = np.polyfit(y_train, y_nn_train_pred, 1)
p = np.poly1d(z)
plt.plot(y_train,p(y_train),"#2c8c2b", label="RF")
ax.set(xlabel='Observed ' + indCol, ylabel='Predicted ' + indCol)
plt.legend(loc="upper left")
ax.title.set_text('Random Forest Result')
return y_nn_train_pred, nn_results
#Wrapper function goes here!
def multiTraining(df,tSize,mList,indCol, nRows, nCols):
"""
Args:
df: (df) Pandas dataframe containing data for training
tSize: (int) Value indicating splitting of data into test and train sets.
Must be between 0 and 1.
mList (str) List of Regressor Models to try. List needs to be defined using at least
one of the following acronyms:
- LR , RF , GP , HR , KNN , NN
indCol (str) Name of column in dataframe that contains the independent variable
nRows (int) How many rows to distribute plots into
nCols (int) How many columns to distribute plots into
"""
#Initialize arrays
listOfPredYVals = []
hexColorList = []
labelList = []
titleList = []
resultList = []
if ("LR" in mList):
print("Building model for Linear Regression")
y_lr_train_pred, lr_results = ML_LRModelTrain(df,tSize,indCol, plotResults = False)
listOfPredYVals.append(y_lr_train_pred)
hexColorList.append("#415c90")
labelList.append("LR")
titleList.append('Linear Regression Result')
resultList.append(lr_results)
if ("RF" in mList):
print("Building model for Linear Regression")
y_rf_train_pred, rf_results = ML_RFModelTrain(df,tSize,indCol, plotResults = False)
listOfPredYVals.append(y_lr_train_pred)
hexColorList.append("#ac4228")
labelList.append("RF")
titleList.append('Random Forest Result')
resultList.append(rf_results)
if ("GP" in mList):
print("Building model for Linear Regression")
y_gp_train_pred, gp_results = ML_GPModelTrain(df,tSize,indCol, plotResults = False)
listOfPredYVals.append(y_lr_train_pred)
hexColorList.append("#28812e")
labelList.append("GP")
titleList.append('Gaussian Process Result')
resultList.append(gp_results)
if ("HR" in mList):
print("Building model for Linear Regression")
y_hp_train_pred, hr_results = ML_HPModelTrain(df,tSize,indCol, plotResults = False)
listOfPredYVals.append(y_lr_train_pred)
hexColorList.append("#581845")
labelList.append("HR")
titleList.append('Huber Process Result')
resultList.append(hr_results)
if ("KNN" in mList):
print("Building model for Linear Regression")
y_kn_train_pred, kn_results = ML_KNNModelTrain(df,tSize,indCol, plotResults = False)
listOfPredYVals.append(y_lr_train_pred)
hexColorList.append("#FF5733")
labelList.append("KNN")
titleList.append('K-Nearest Neighbors Result')
resultList.append(kn_results)
if ("NN" in mList):
print("Building model for Linear Regression")
y_nn_train_pred, nn_results = ML_NNModelTrain(df,tSize,indCol, plotResults = False)
listOfPredYVals.append(y_lr_train_pred)
hexColorList.append("#FFC300")
labelList.append("NN")
titleList.append('Neural Network Result')
resultList.append(nn_results)
#Combine results from all our models into a single dataframe
mod_Results = pd.concat(resultList,ignore_index=True).sort_values('Training R2',
axis = 0, ascending = False)
mod_Results
#Plot the results
multiMLModelPlotting(listOfPredYVals, hexColorList, labelList, titleList, nRows, nCols, mod_Results)
return 0
#Plotting function for multi model routine --> There are some bugs in it still
def multiMLModelPlotting(listOfPredYVals, hexColorList, labelList, titleList, nRows, nCols, mod_Results):
N = len(titleList)
#Visualize results of linear regression
plt.rcParams.update({'font.size': 20})
pltRows = nRows #
pltCols = nCols #
delPlot = False
fig, axs = plt.subplots(nrows = pltRows, ncols = pltCols, figsize=(15,15) , squeeze = False)
i = 0; j = 0; k = 0;
for element in listOfPredYVals:
color = hexColorList[k]
yTrainPredPlot = listOfPredYVals[k]
label = labelList[k]
#print(i, j, label, k)
#Make scatter plot of train set and regressor model
axs[i,j].scatter(x = y_train, y = yTrainPredPlot, c=color, alpha=0.5)
#Fit a first order polynomial (i.e. a straight line) to the regressor model
z = np.polyfit(y_train, yTrainPredPlot, 1)
p = np.poly1d(z)
#Add labels and colors and stuff
val = mod_Results.loc[k,'Training R2'] #Get the r2 value from the model results dataframe
val = "{:.2f}".format(val)
axs[i,j].plot(y_train,p(y_train),"#b20cd7", label=label+"\nr\u00b2".format(2) + " = " + str(val))
axs[i,j].title.set_text(titleList[k])
axs[i,j].legend(loc="upper left")
axs[i,j].grid(color = 'black', linestyle = '--', linewidth = 0.5)
axs[i,j].set(xlabel='Experimental LogS', ylabel='Predicted LogS')
axs[i,j].label_outer()
if i < pltRows - 1:
i += 1
else:
i = 0
j += 1
if (k+1 >= N) & ((nRows * nCols) >= N):
fig.delaxes(axs[i][j])
if (k >= N) & ((nRows * nCols) >= N):
fig.delaxes(axs[nRows-1][nCols-1])
k += 1
plt.show()